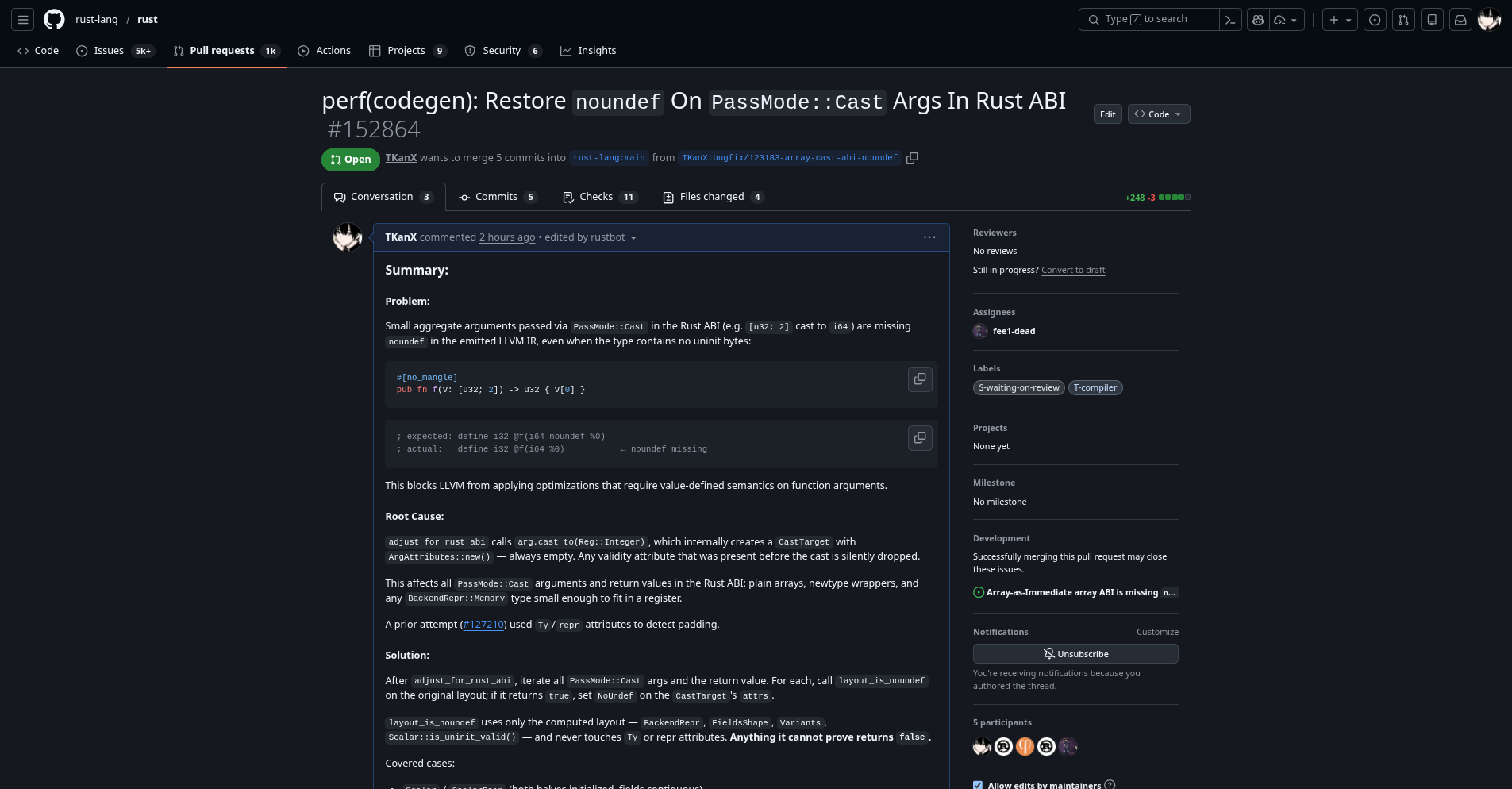
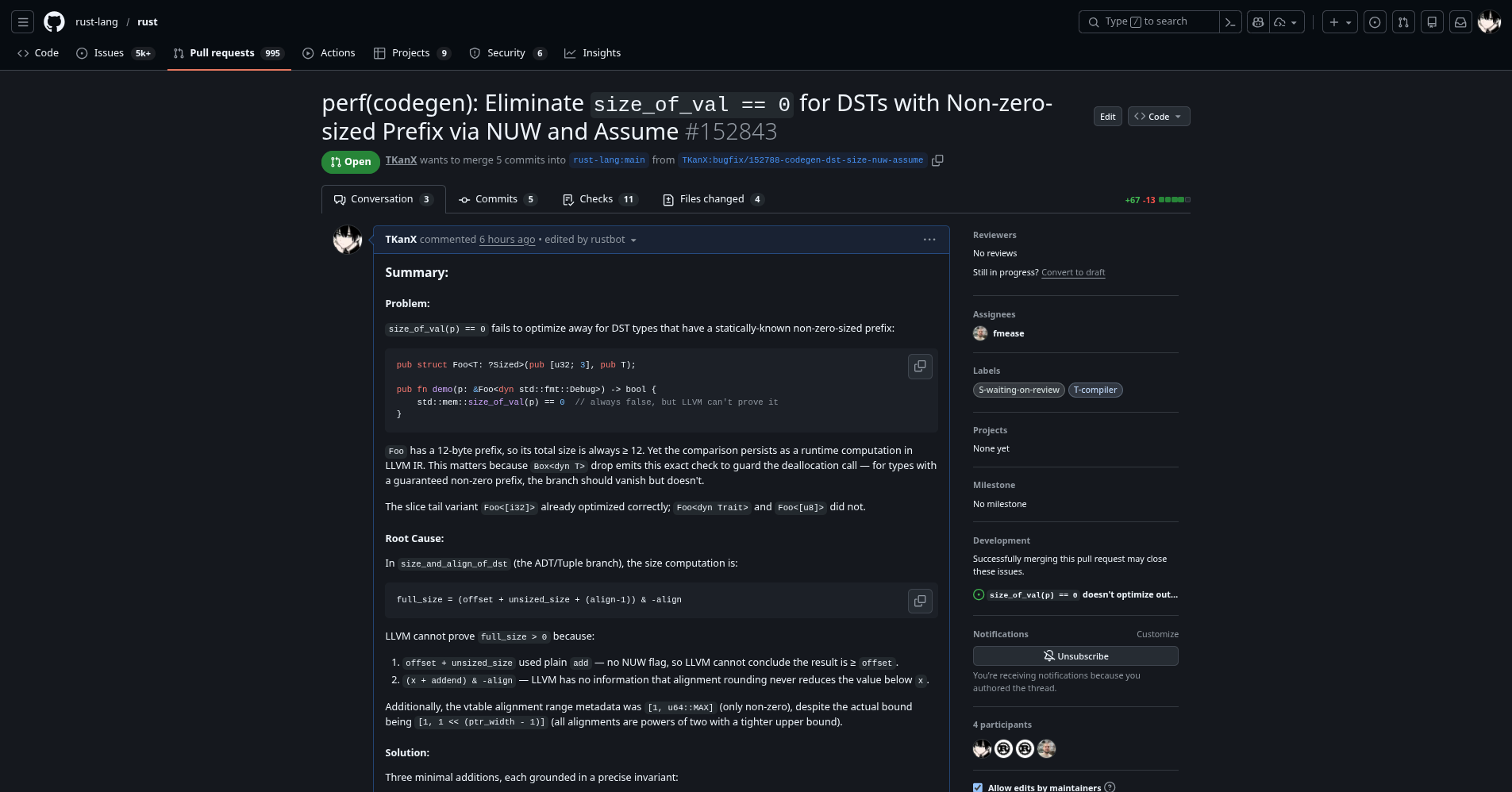
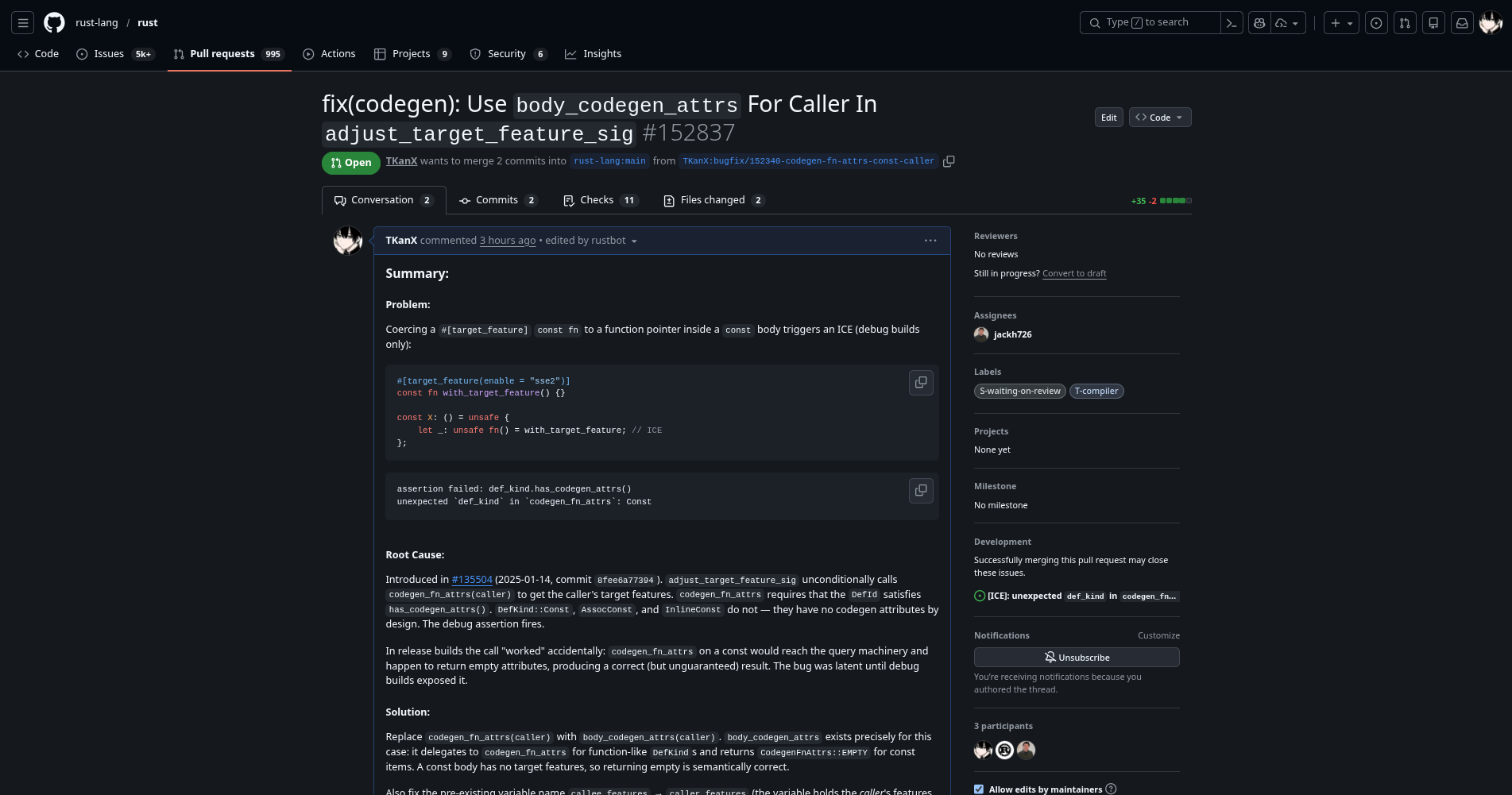
Optimized the Performance of LLVM Generated by the Compiler
https://github.com/rust-lang/rust/pull/152864
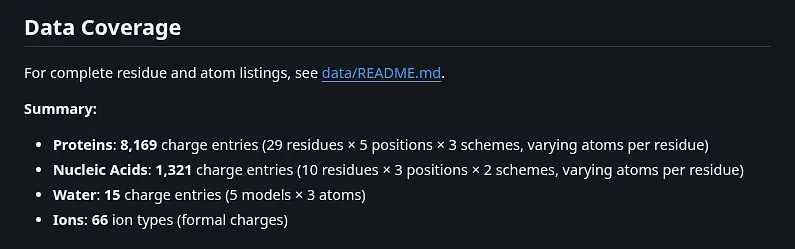
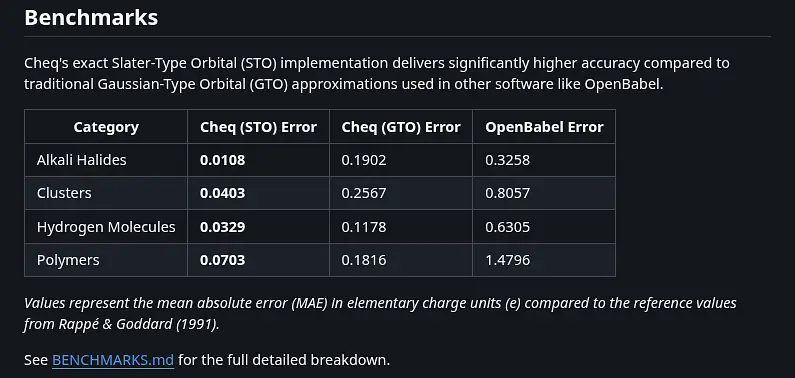
Summary:
Problem:
Small aggregate arguments passed via PassMode::Cast in the Rust ABI (e.g. [u32; 2] cast to i64) are missing noundef in the emitted LLVM IR, even when the type contains no uninit bytes:
#[no_mangle]
pub fn f(v: [u32; 2]) -> u32 { v[0] }
; expected: define i32 @f(i64 noundef %0)
; actual: define i32 @f(i64 %0) ← noundef missing
This blocks LLVM from applying optimizations that require value-defined semantics on function arguments.
Root Cause:
adjust_for_rust_abi calls arg.cast_to(Reg::Integer), which internally creates a CastTarget with ArgAttributes::new() — always empty. Any validity attribute that was present before the cast is silently dropped.
This affects all PassMode::Cast arguments and return values in the Rust ABI: plain arrays, newtype wrappers, and any BackendRepr::Memory type small enough to fit in a register.
A prior attempt (rust-lang/rust#127210) used Ty/repr attributes to detect padding.
Solution:
After adjust_for_rust_abi, iterate all PassMode::Cast args and the return value. For each, call layout_is_noundef on the original layout; if it returns true, set NoUndef on the CastTarget’s attrs.
layout_is_noundef uses only the computed layout — BackendRepr, FieldsShape, Variants, Scalar::is_uninit_valid() — and never touches Ty or repr attributes. Anything it cannot prove returns false.
Covered cases:
-
Scalar/ScalarPair(both halves initialized, fields contiguous) -
FieldsShape::Array(element type recursively uninit-free) -
FieldsShape::ArbitrarywithVariants::Single(fields cover0..sizewith no gaps, each recursively uninit-free) — handles newtype wrappers, multi-field structs, single-variant enums,repr(transparent),repr(C)wrappers
Log in to leave a comment





.jpg)
%20(1).jpg)
%20(2).jpg)