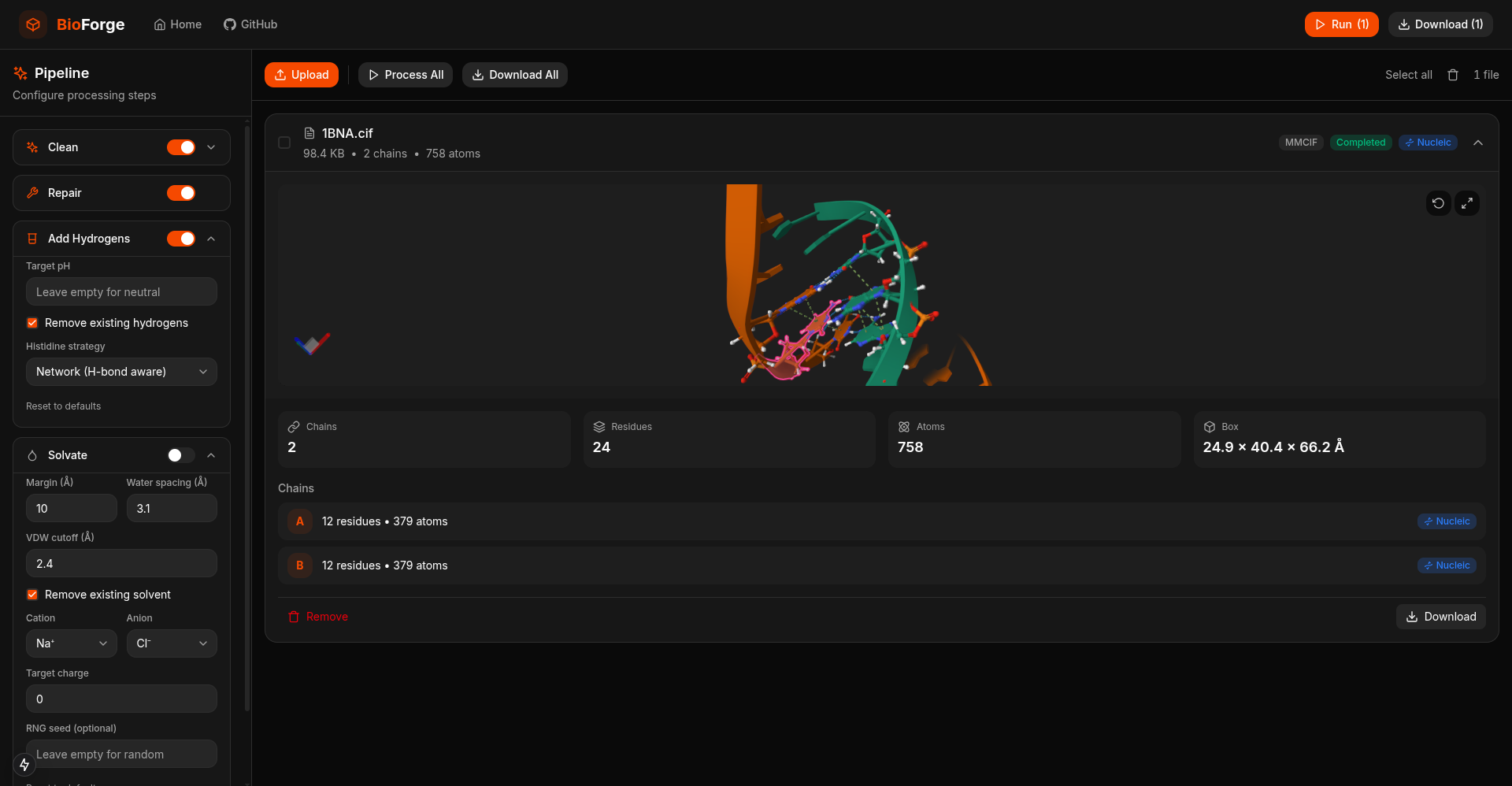
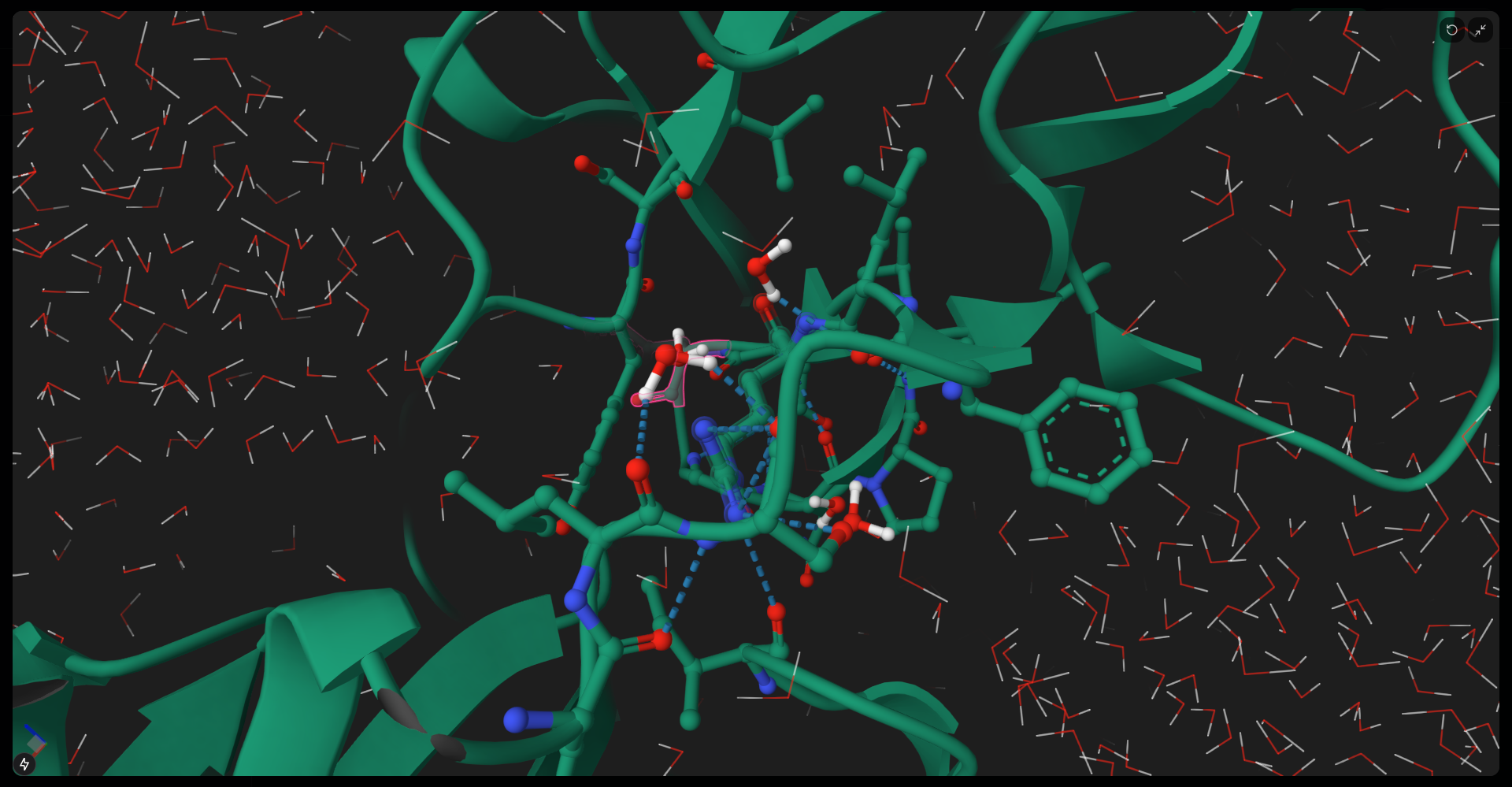
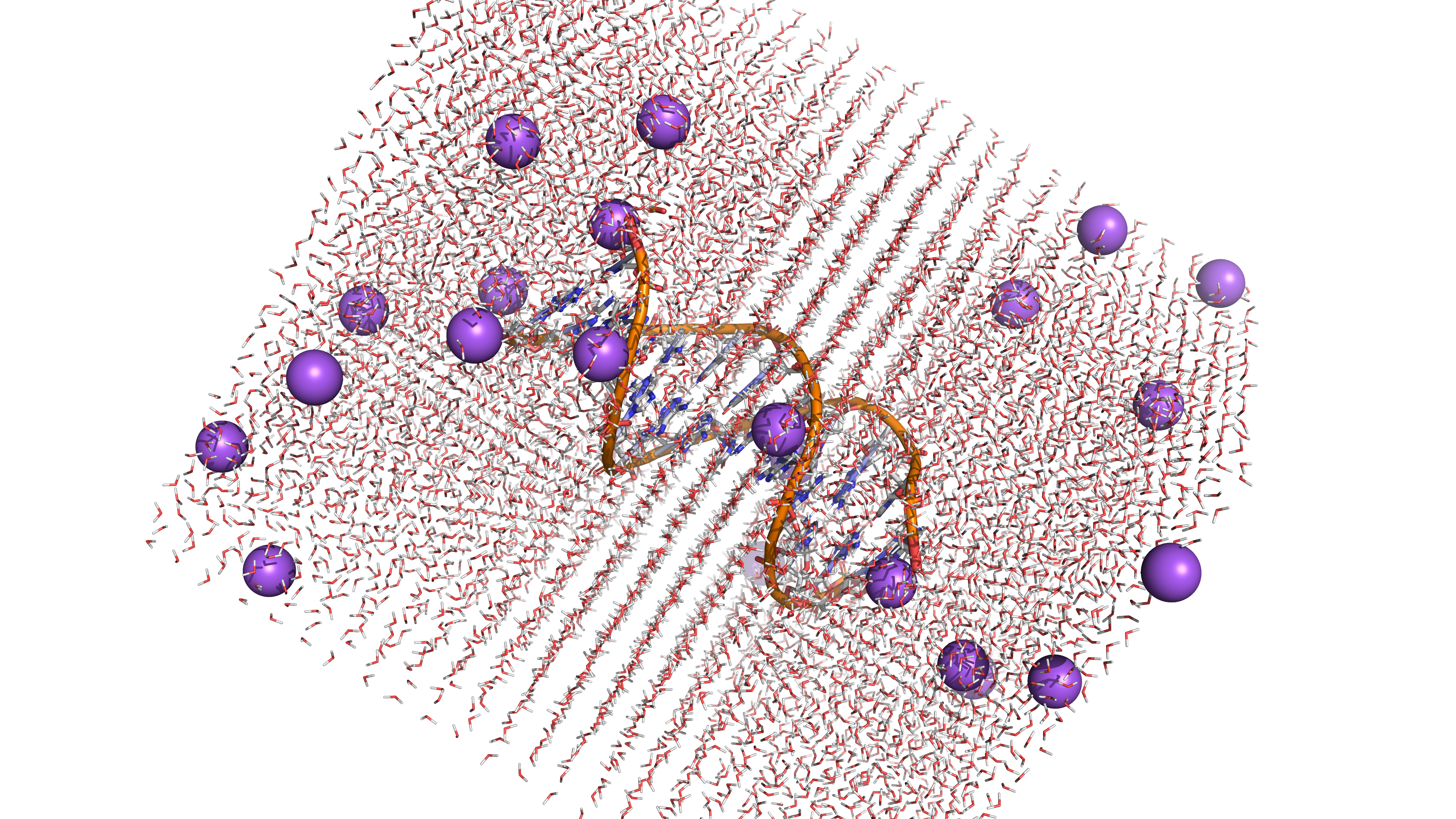
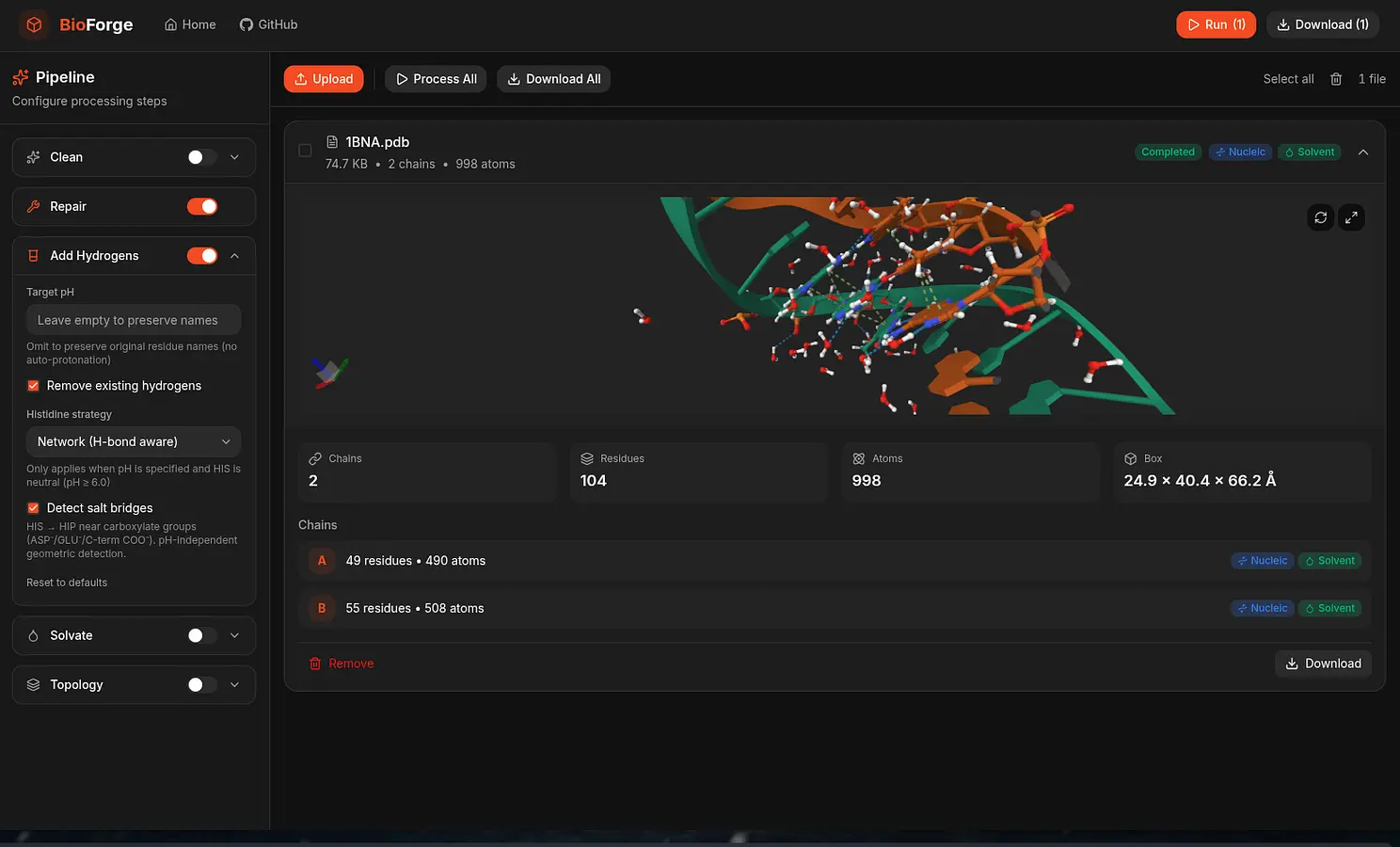
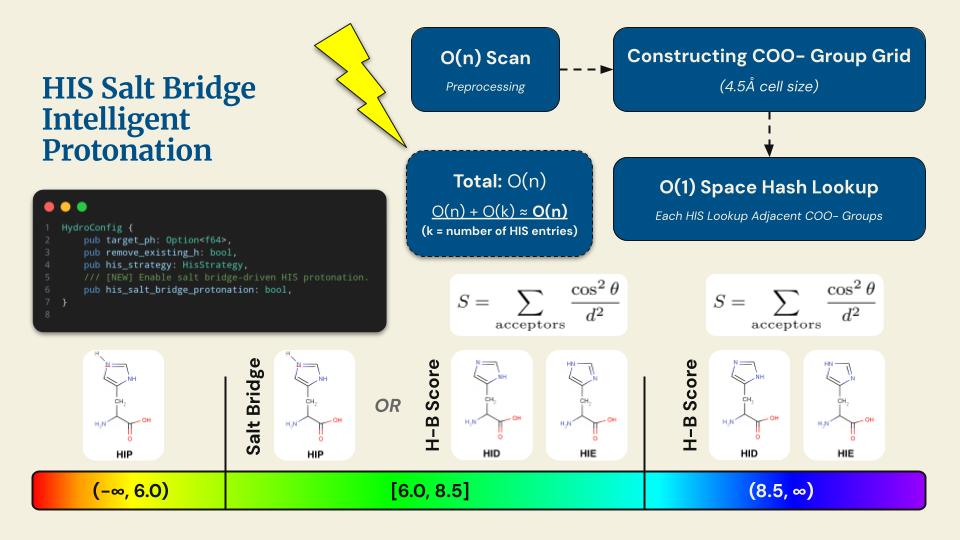
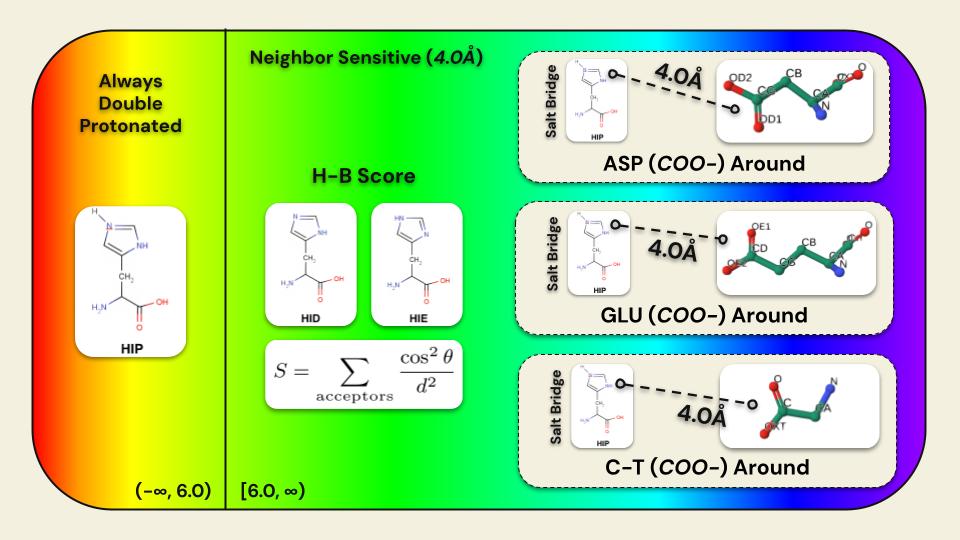
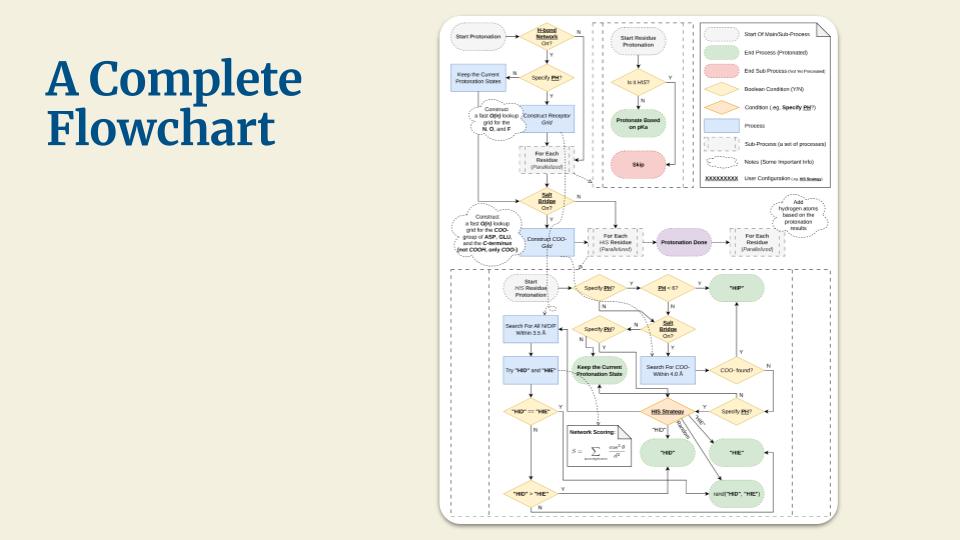
A high-performance, pure-Rust toolkit for standardizing and preparing DNA, RNA, and protein biomolecular systems. It repairs missing atoms, assigns protonation states, adds solvation layers, and unifies topologies to generate simulation-ready structures. The toolkit can process entire viral capsids at millisecond scale, and is available as a Web application, CLI, and reusable library, with built-in parallelization and spatial indexing for maximum performance and scalability.
(Viruses of millions atoms => takes only 1s)

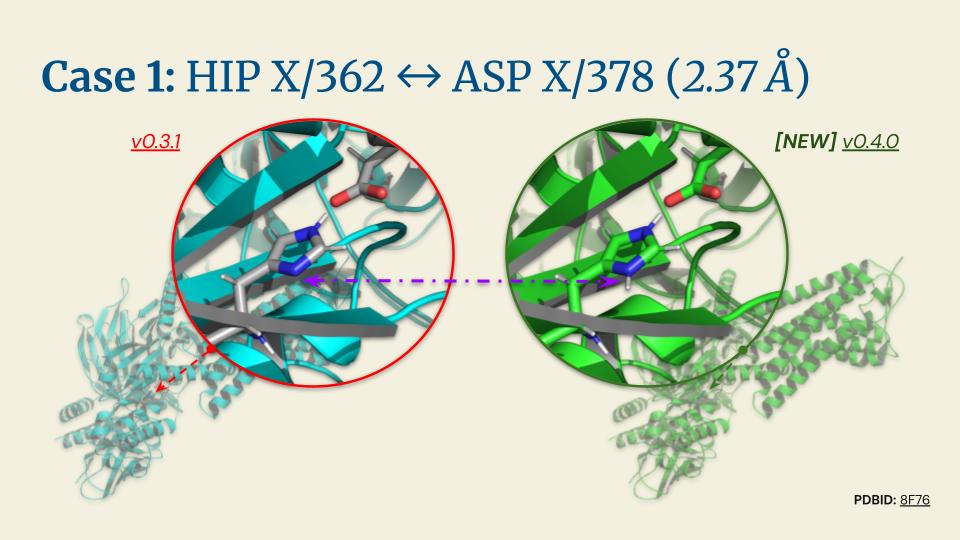
.jpg)
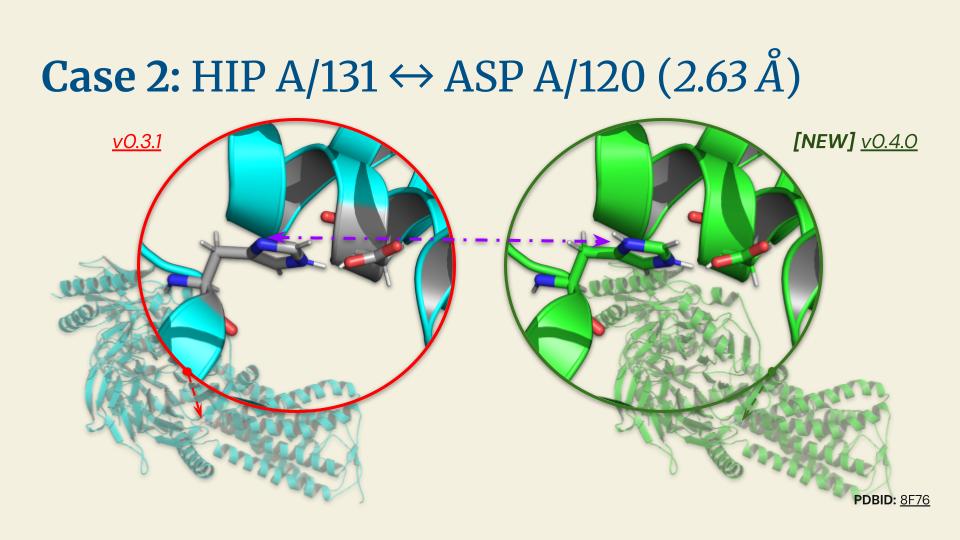
%20(1).jpg)
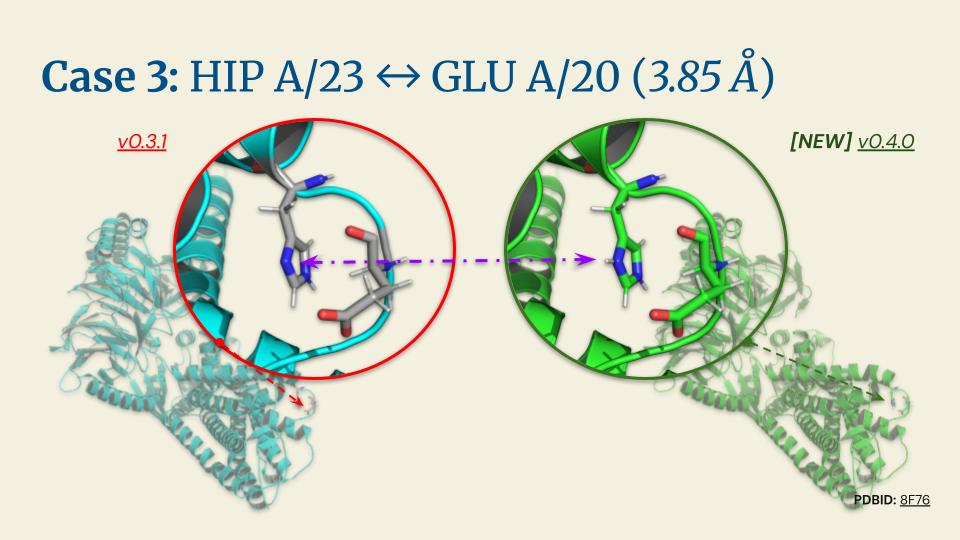
%20(2).jpg)